sandbox/acastillo/output_fields/vtkhdf/output_vtkhdf.h
VTKHDF File Format
These routines are compatible with the
VTKHDF Model and Format which can be read using Paraview or
Visit.
Data is composed of large datasets stored using the Hierarchical Data
Format HDF5. As the name implies, data is organized following a
hierarchical structure. HDF5 files can be read without any prior
knowledge of the stored data. The type, rank, dimension and other
properties of each array are stored inside the file in the form of
meta-data. Additional features include support for a large number of
objects, file compression, a parallel I/O implementation through the
MPI-IO or MPI POSIX drivers. Using this format requires the HDF5
library, which is usually installed in most computing centers or may be
installed locally through a repository. Linking is automatic but
requires the environment variables HDF5_INCDIR and
HDF5_LIBDIR, which are usually set when you load the module
hdf5. You might also add something like this to your
Makefile
HDFLIBS=-I$(HDF5_INCDIR) -L$(HDF5_LIBDIR) -lhdf5 -lhdf5_hl
LIBS+=$(HDFLIBS)To use HDF5 we need to “declare” the library in the qcc
system. This should not be the case, but HDF5 is a messy
library that just won’t pass through qcc
echo "@include <hdf5.h>" > $BASILISK/ast/std/hdf5.h
echo "@include <hdf5_hl.h>" > $BASILISK/ast/std/hdf5_hl.hWe also need to declare the datatypes at the end of
$BASILISK/ast/defaults.h
echo "typedef hid_t, hsize_t, herr_t, H5L_info_t;" >> $BASILISK/ast/defaults.hAn explanation can be found here
#pragma autolink -lhdf5
#include <hdf5.h>preamble: define some useful macros
#define shortcut_slice(n, _alpha) \
double alpha = (_alpha - n.x * x - n.y * y - n.z * z) / Delta; \
if (fabs(alpha) > 0.87) \
continue;
#include "output_vtkhdf_helpers.h"output_vtkhdf(): Exports full 2D (or 3D) fields.
This function writes one VTKHDF
file which can be read using Paraview. The VTKHDF file format is a
file format relying on HDF5. It is meant to provide good I/O performance
as well as robust and flexible parallel I/O capabilities. The file
stores scalar and vector fields defined at the center points are stored
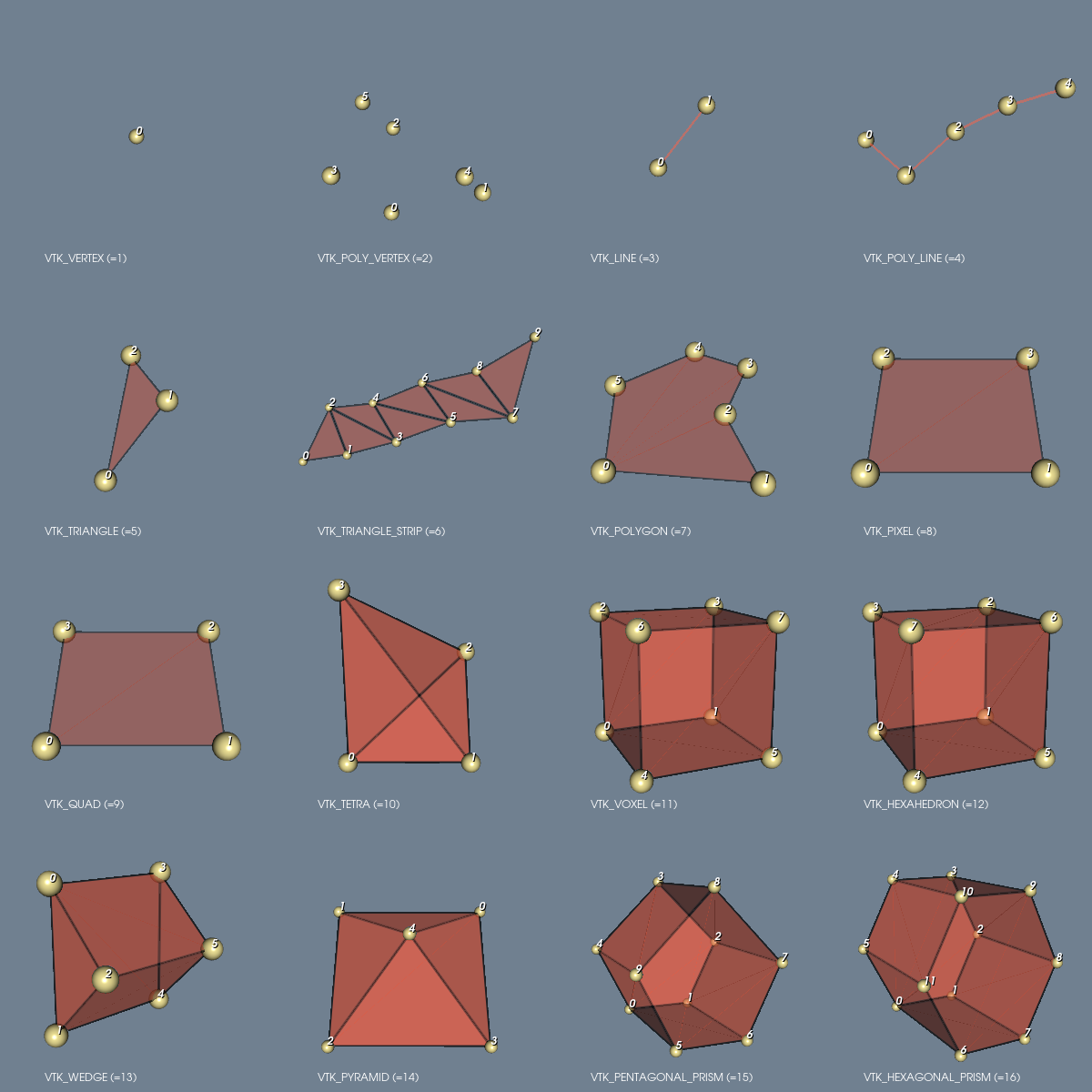
at the cell center of an unstructured grid with type
VTK_QUAD in 2D, or VTK_HEXAHEDRON in 3D.
A quick description of the file format is copied from here.
VTK HDF files start with a group called VTKHDF with two
attributes: Version, an array of two integers and
Type, a string showing the VTK dataset type stored in the
file — in our case UnstructuredGrid. The data type for each
HDF dataset is part of the dataset and it is determined at write time.
In the diagram below, showing the HDF file structure for
UnstructuredGrid, the rounded blue rectangles are HDF
groups and the gray rectangles are HDF datasets. Each rectangle shows
the name of the group or dataset in bold font and the attributes
underneath with regular font. In our case, scalar and
vector fields are stored as CellData. The
unstructured grid is split into partitions, with a partition for each
MPI rank.
|
|
|
|
|
|
The arguments and their default values are:
- slist : Pointer to an array of scalar data.
- vlist : Pointer to an array of vector data.
- name : Output file name generally uses the
.vtkhdfextension. - compression_level : Level of compression to use when writing data to the HDF5 file (default=9).
Example Usage
scalar * slist = {a,b};
vector * vlist = {c,d};
output_vtkhdf(slist, vlist, "domain.vtkhdf");trace void output_vtkhdf(scalar *slist, vector *vlist, char *name = "domain.vtkhdf", int compression_level = 9){
hid_t acc_tpl1; // File access template
hid_t file_id; // HDF5 file ID
hid_t group_id; // HDF5 group ID
hid_t subgroup_id; // HDF5 subgroup ID
hsize_t count[2]; // Hyperslab selection parameters
hsize_t offset[2]; // Offset for hyperslab
hsize_t dims[1] = {2};
// Define a scalar mask for periodic conditions
scalar per_mask[];
foreach () {
per_mask[] = 1.;
}
// VTK cell types: VTK_QUAD (in 2D) or VTK_HEXAHEDRON (in 3D)
int type, noffset;
#if dimension == 2
type = 9;
noffset = 4;
#elif dimension == 3
type = 12;
noffset = 8;
#endif
// Obtain the number of points and cells and get a marker to reconstruct the topology
int number_of_points = 0, number_of_points_loc = 0;
int number_of_cells = 0, number_of_cells_loc = 0;
count_points_and_cells(&number_of_points, &number_of_cells, &number_of_points_loc, &number_of_cells_loc, per_mask);
int number_of_ids = number_of_cells*noffset;
int number_of_ids_loc = number_of_cells_loc*noffset;
// Define chunk size for parallel I/O
hsize_t chunk_size = number_of_cells / npe() / 8;
// Calculate offsets for parallel I/O
int offset_points[npe()], offset_cells[npe()], offset_ids[npe()], offset_offset[npe()];
calculate_offsets2(offset_offset, number_of_cells_loc+1, offset);
calculate_offsets2(offset_ids, number_of_ids_loc, offset);
calculate_offsets2(offset_cells, number_of_cells_loc, offset);
calculate_offsets2(offset_points, number_of_points_loc, offset);
// Initialize marker for topology reconstruction
vertex scalar marker[];
initialize_marker(marker, offset, 0);
// Setup file access template with parallel I/O access
acc_tpl1 = H5Pcreate(H5P_FILE_ACCESS);
H5Pset_fapl_mpio(acc_tpl1, MPI_COMM_WORLD, MPI_INFO_NULL);
// Create a new HDF5 file collectively
file_id = H5Fcreate(name, H5F_ACC_TRUNC, H5P_DEFAULT, acc_tpl1);
// Release file-access template
H5Pclose(acc_tpl1);
// Create group
group_id = H5Gcreate(file_id, "VTKHDF", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT);
// Create "Version", "Type" (and other) attributes
dims[0] = 2;
int version_data[2] = {2, 1};
create_attribute(group_id, "Version", version_data, dims);
create_attribute_type(group_id, "Type", "UnstructuredGrid", 16);
create_attribute_type(group_id, "Description", "Simulation perfomed using Basilisk (http://basilisk.fr/)", 57);
// Write "NumberOfConnectivityIds", "NumberOfPoints", "NumberOfCells"
dims[0] = npe();
write_simple_dataset(group_id, "NumberOfConnectivityIds", offset_ids, dims);
write_simple_dataset(group_id, "NumberOfPoints", offset_points, dims);
write_simple_dataset(group_id, "NumberOfCells", offset_cells, dims);
// Populate and write the points dataset
double *points_dset;
populate_points_dset(&points_dset, number_of_points_loc, offset_points, count, offset);
create_chunked_dataset(group_id, count, offset, "Points", number_of_points, number_of_points_loc, 3, points_dset, H5T_NATIVE_DOUBLE, chunk_size, compression_level);
free(points_dset);
// Populate and write the types dataset
char * types_dset;
populate_types_dset(&types_dset, type, number_of_cells_loc, offset_cells, count, offset);
create_chunked_dataset(group_id, count, offset, "Types", number_of_cells, number_of_cells_loc, 1, types_dset, H5T_STD_U8LE, chunk_size, compression_level);
free(types_dset);
// Populate and write the connectivity dataset
long *topo_dset;
populate_topo_dset(&topo_dset, number_of_cells_loc, offset_ids, count, offset, per_mask, marker);
create_chunked_dataset(group_id, count, offset, "Connectivity", number_of_ids, number_of_ids_loc, 1, topo_dset, H5T_NATIVE_LONG, chunk_size, compression_level);
free(topo_dset);
// Populate and write the offsets dataset
long *offsets_dset;
populate_offsets_dset(&offsets_dset, noffset, number_of_cells_loc+1, offset_offset, count, offset);
create_chunked_dataset(group_id, count, offset, "Offsets", number_of_cells+npe(), number_of_cells_loc+1, 1, offsets_dset, H5T_NATIVE_LONG, chunk_size, compression_level);
free(offsets_dset);
// Create subgroup "CellData"
subgroup_id = H5Gcreate(group_id, "CellData", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT);
// Allocate memory and write scalar datasets
double *scalar_dset = (double *)malloc(number_of_cells_loc * sizeof(double));
for (scalar s in slist) {
populate_scalar_dset(s, scalar_dset, number_of_cells_loc, offset_cells, count, offset, per_mask);
create_chunked_dataset(subgroup_id, count, offset, s.name, number_of_cells, number_of_cells_loc, 1, scalar_dset, H5T_NATIVE_DOUBLE, chunk_size, compression_level);
}
free(scalar_dset);
// Allocate memory and write vector datasets
double *vector_dset = (double *)malloc(number_of_cells_loc * 3 * sizeof(double));
for (vector v in vlist) {
populate_vector_dset(v, vector_dset, number_of_cells_loc, offset_cells, count, offset, per_mask);
create_chunked_dataset(subgroup_id, count, offset, v.x.name, number_of_cells, number_of_cells_loc, 3, vector_dset, H5T_NATIVE_DOUBLE, chunk_size, compression_level);
}
free(vector_dset);
H5Gclose(subgroup_id);
// Create subgroup "FieldData"
subgroup_id = H5Gcreate(group_id, "FieldData", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT);
H5Gclose(subgroup_id);
// Create subgroup "PointData"
subgroup_id = H5Gcreate(group_id, "PointData", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT);
H5Gclose(subgroup_id);
H5Gclose(group_id);
// Close HDF5 resources
H5Fclose(file_id);
}output_vtkhdf_slice(): Exports a 2D slice of a 3D field along x, y, or z.
This function takes a slice defined by n={n_x, n_y, n_z}
and _alpha as as in view.h. Only works
for x, y, and/or z planes. Here,
_alpha is assumed to intersect a cell face and
must be a multiple of
L0/1<<MINLEVEL. This also means that scalar and
vector fields are written at the corresponding face values.
Naturaly, this routine only works in 3D.
The arguments and their default values are:
- list
- pointer to the list of scalar fields to be exported.
- vlist
- pointer to the list of vector fields to be exported.
- name
-
Output file name generally uses the
.vtkhdfextension. - n
- vector \vec{n} normal to the plane.
- alpha
- coordinate \alpha intersecting the plane.
- compression_level
- Level of compression to use when writing data to the HDF5 file (default=9).
Example Usage
scalar * slist = {a,b};
vector * vlist = {c,d};
output_vtkhdf_slice(slist, vlist, "slice_x.vtkhdf", (coord){1,0,0}, 0);
output_vtkhdf_slice(slist, vlist, "slice_y.vtkhdf", (coord){0,1,0}, 0);
output_vtkhdf_slice(slist, vlist, "slice_z.vtkhdf", (coord){0,0,1}, 0);see, also example.
#if dimension == 3
trace void output_vtkhdf_slice(scalar *slist, vector *vlist, char *name, coord n = {0, 0, 1}, double _alpha = 0, int compression_level = 9){
hid_t acc_tpl1; // File access template
hid_t file_id; // HDF5 file ID
hid_t group_id; // HDF5 group ID
hid_t subgroup_id; // HDF5 subgroup ID
hsize_t count[2]; // Hyperslab selection parameters
hsize_t offset[2]; // Offset for hyperslab
hsize_t dims[1] = {2};
// Define a scalar mask to deal with solids and periodic conditions
scalar per_mask[];
foreach (){
per_mask[] = 0.;
shortcut_slice(n, _alpha);
if (alpha > 0.){
#if EMBED
per_mask[] = cs[];
#else
per_mask[] = 1.;
#endif
}
}
// VTK cell types: VTK_QUAD
int type = 9;
int noffset = 4;
// Obtain the number of points and cells and get a marker to reconstruct the topology
int number_of_points = 0, number_of_points_loc = 0;
int number_of_cells = 0, number_of_cells_loc = 0;
count_points_and_cells_slice(&number_of_points, &number_of_cells, &number_of_points_loc, &number_of_cells_loc, per_mask, n, _alpha);
int number_of_ids = number_of_cells*noffset;
int number_of_ids_loc = number_of_cells_loc*noffset;
// Define chunk size for parallel I/O
hsize_t chunk_size = number_of_cells / npe() / 2;
// Calculate offsets for parallel I/O
int offset_points[npe()], offset_cells[npe()], offset_ids[npe()], offset_offset[npe()];
calculate_offsets2(offset_offset, number_of_cells_loc+1, offset);
calculate_offsets2(offset_ids, number_of_ids_loc, offset);
calculate_offsets2(offset_cells, number_of_cells_loc, offset);
calculate_offsets2(offset_points, number_of_points_loc, offset);
// Initialize marker for topology reconstruction
vertex scalar marker[];
initialize_marker_slice(marker, offset, n, _alpha, 0);
// Setup file access template with parallel I/O access
acc_tpl1 = H5Pcreate(H5P_FILE_ACCESS);
H5Pset_fapl_mpio(acc_tpl1, MPI_COMM_WORLD, MPI_INFO_NULL);
// Create a new HDF5 file collectively
file_id = H5Fcreate(name, H5F_ACC_TRUNC, H5P_DEFAULT, acc_tpl1);
// Release file-access template
H5Pclose(acc_tpl1);
// Create group
group_id = H5Gcreate(file_id, "VTKHDF", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT);
// Create "Version", "Type" (and other) attributes
dims[0] = 2;
int version_data[2] = {2, 1};
create_attribute(group_id, "Version", version_data, dims);
create_attribute_type(group_id, "Type", "UnstructuredGrid", 16);
create_attribute_type(group_id, "Description", "Simulation perfomed using Basilisk (http://basilisk.fr/)", 57);
// Write "NumberOfConnectivityIds", "NumberOfPoints", "NumberOfCells"
dims[0] = npe();
write_simple_dataset(group_id, "NumberOfConnectivityIds", offset_ids, dims);
write_simple_dataset(group_id, "NumberOfPoints", offset_points, dims);
write_simple_dataset(group_id, "NumberOfCells", offset_cells, dims);
// Populate and write the points dataset
double *points_dset;
populate_points_dset_slice(&points_dset, number_of_points_loc, offset_points, count, offset, n, _alpha);
create_chunked_dataset(group_id, count, offset, "Points", number_of_points, number_of_points_loc, 3, points_dset, H5T_NATIVE_DOUBLE, chunk_size, compression_level);
free(points_dset);
// Populate and write the types dataset
char * types_dset;
populate_types_dset(&types_dset, type, number_of_cells_loc, offset_cells, count, offset);
create_chunked_dataset(group_id, count, offset, "Types", number_of_cells, number_of_cells_loc, 1, types_dset, H5T_STD_U8LE, chunk_size, compression_level);
free(types_dset);
// Populate and write the connectivity dataset
long *topo_dset;
populate_topo_dset_slice(&topo_dset, number_of_cells_loc, offset_ids, count, offset, per_mask, marker, n, _alpha);
create_chunked_dataset(group_id, count, offset, "Connectivity", number_of_ids, number_of_ids_loc, 1, topo_dset, H5T_NATIVE_LONG, chunk_size, compression_level);
free(topo_dset);
// Populate and write the offsets dataset
long *offsets_dset;
populate_offsets_dset(&offsets_dset, noffset, number_of_cells_loc+1, offset_offset, count, offset);
create_chunked_dataset(group_id, count, offset, "Offsets", number_of_cells+npe(), number_of_cells_loc+1, 1, offsets_dset, H5T_NATIVE_LONG, chunk_size, compression_level);
free(offsets_dset);
// Create subgroup "CellData"
subgroup_id = H5Gcreate(group_id, "CellData", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT);
// Allocate memory and write scalar datasets
double *scalar_dset = (double *)malloc(number_of_cells_loc * sizeof(double));
for (scalar s in slist) {
populate_scalar_dset_slice(s, scalar_dset, number_of_cells_loc, offset_cells, count, offset, per_mask, n, _alpha);
create_chunked_dataset(subgroup_id, count, offset, s.name, number_of_cells, number_of_cells_loc, 1, scalar_dset, H5T_NATIVE_DOUBLE, chunk_size, compression_level);
}
free(scalar_dset);
// Allocate memory and write vector datasets
double *vector_dset = (double *)malloc(number_of_cells_loc * 3 * sizeof(double));
for (vector v in vlist) {
populate_vector_dset_slice(v, vector_dset, number_of_cells_loc, offset_cells, count, offset, per_mask, n, _alpha);
create_chunked_dataset(subgroup_id, count, offset, v.x.name, number_of_cells, number_of_cells_loc, 3, vector_dset, H5T_NATIVE_DOUBLE, chunk_size, compression_level);
}
free(vector_dset);
H5Gclose(subgroup_id);
// Create subgroup "FieldData"
subgroup_id = H5Gcreate(group_id, "FieldData", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT);
H5Gclose(subgroup_id);
// Create subgroup "PointData"
subgroup_id = H5Gcreate(group_id, "PointData", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT);
H5Gclose(subgroup_id);
H5Gclose(group_id);
// Close HDF5 resources
H5Fclose(file_id);
}
#endifpostamble: delete macros
#undef shortcut_slice