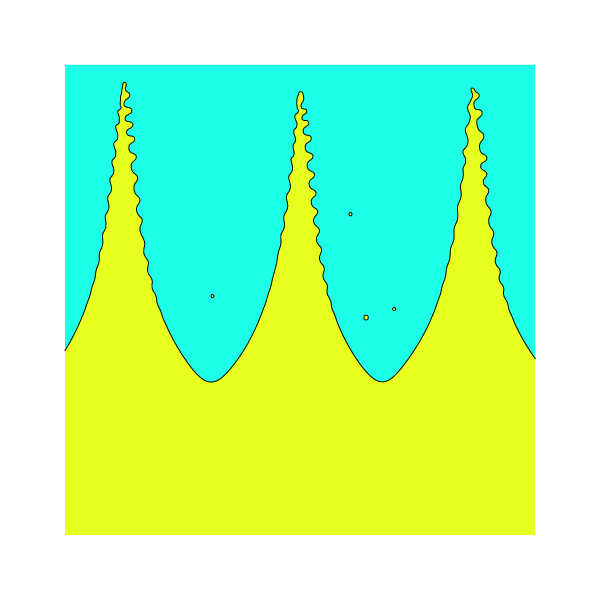
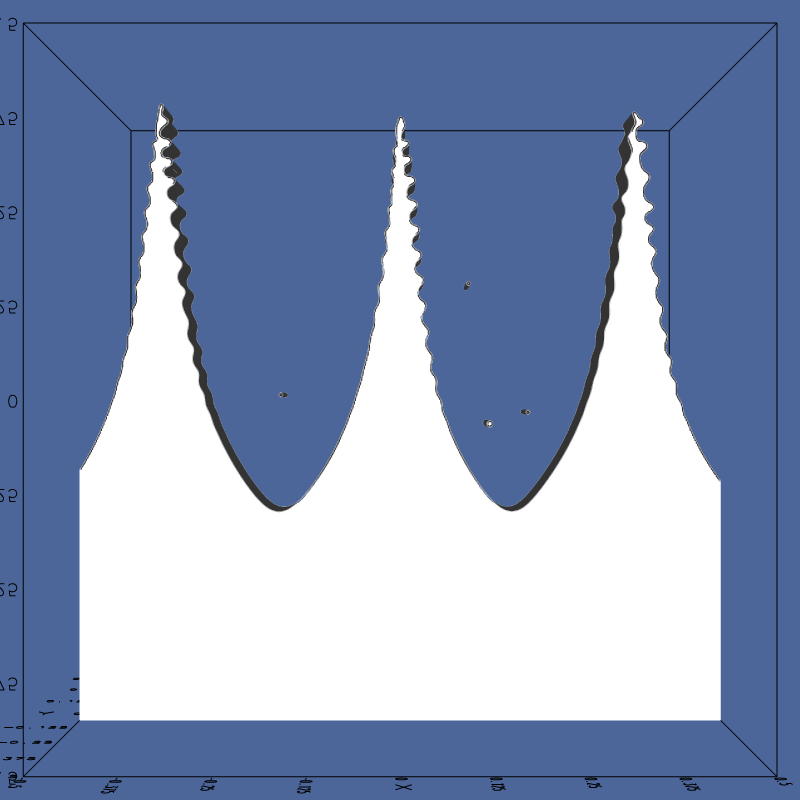
 An example of the intial condition in 2D (left)
and the corresponding 3D interface (right).
An example of the intial condition in 2D (left)
and the corresponding 3D interface (right).

 An example of the intial condition in 2D (left)
and the corresponding 3D interface (right).
An example of the intial condition in 2D (left)
and the corresponding 3D interface (right).
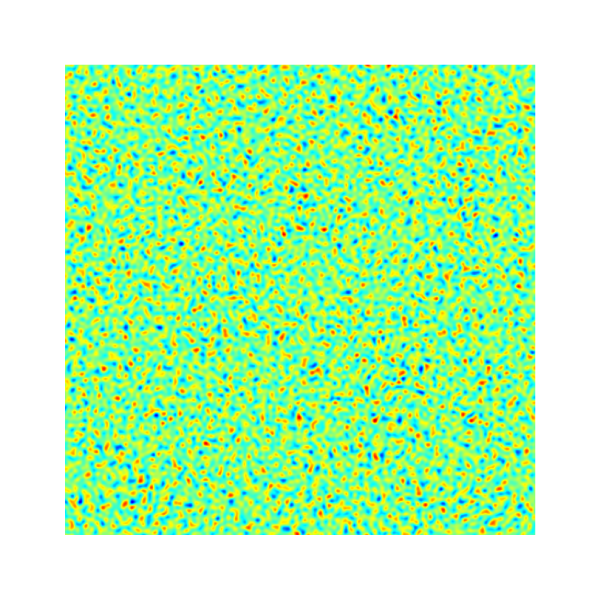
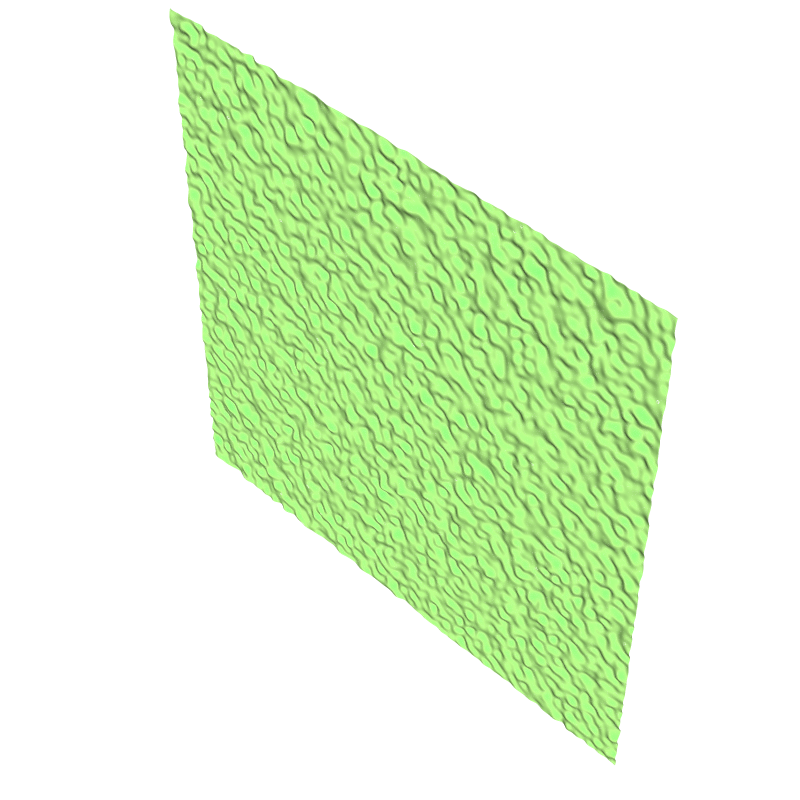 An example of the initialized interface using `isvof=0` (left)
and `isvof=1` (right).
An example of the initialized interface using `isvof=0` (left)
and `isvof=1` (right).